Evolu-sec package
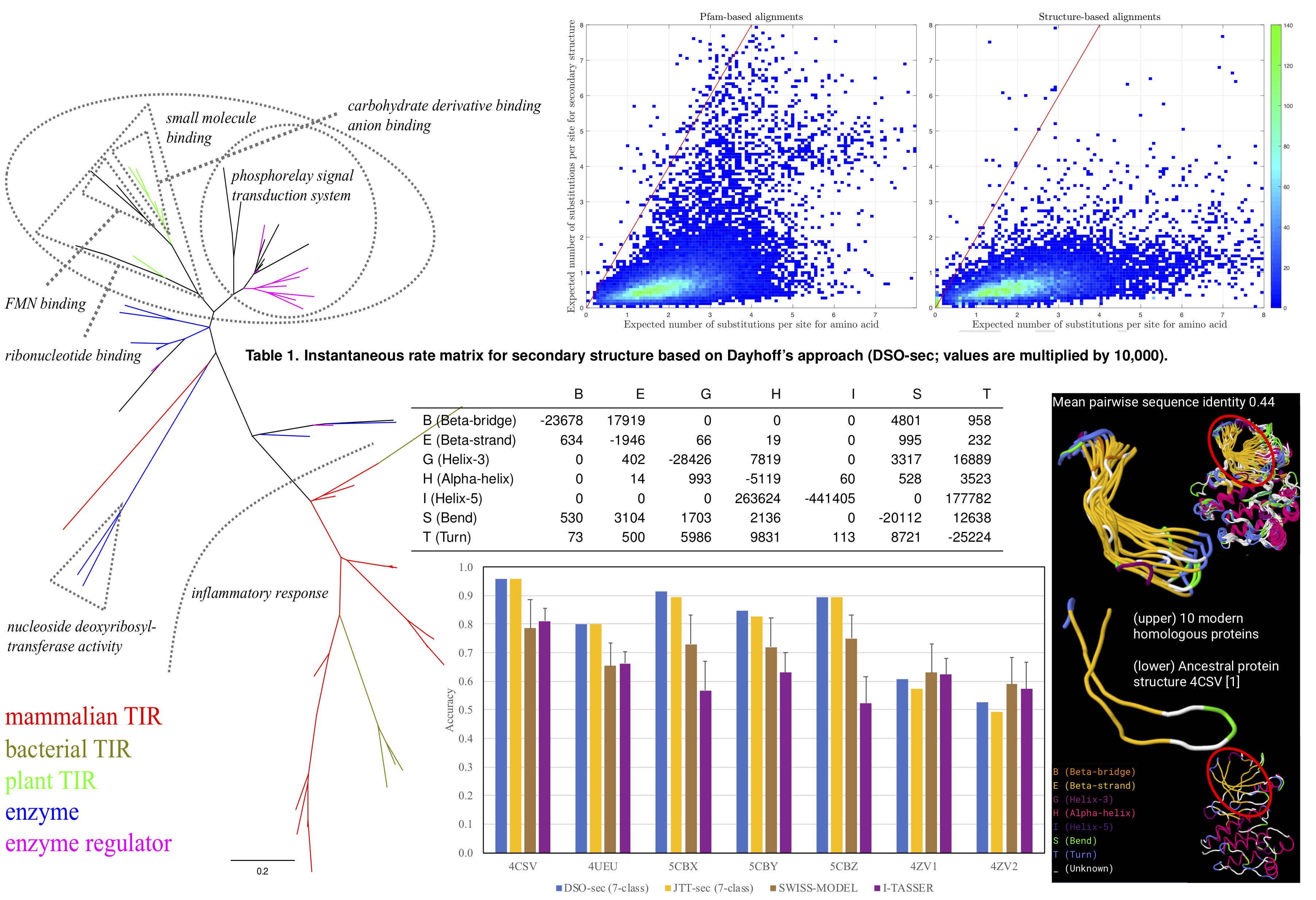
<html> <p class=MsoNormal style='text-align:justify;text-justify:inter-ideograph'><span class=SpellE><span lang=EN-GB>Evolu</span></span><span lang=EN-GB>-sec package forms part of a research project, reported in “Evolutionary model of protein secondary structure capable of revealing new biological relationships”.<o:p></o:p><a href="https://doi.org/10.1002/prot.25898">http://dx.doi.org/10.1101/563452</a></span></p> <p class=MsoNormal style='text-align:justify;text-justify:inter-ideograph'><span class=SpellE><span lang=EN-GB>Evolu</span></span><span lang=EN-GB>-sec package contains phylogenetic analysis tools based on an evolutionary model of secondary structure <b style='mso-bidi-font-weight:normal'>[centre, Table 1]</b>. The analysis focuses on phylogenetic tree inference <b style='mso-bidi-font-weight: normal'>[left]</b>, evolutionary distance estimation <b style='mso-bidi-font-weight: normal'>[top]</b>, and ancestral secondary structure reconstruction. <b style='mso-bidi-font-weight:normal'>[bottom and right]</b>.</span></p> </html>
The research project abstract is provided in the next section. The dataset (140.9MB) for the model development and the software can be accessed in the “Supplementary Data” section.
Research Project
Evolutionary model of protein secondary structure capable of revealing new biological relationships
Jhih-Siang Lai, Burkhard Rost
, Bostjan Kobe and Mikael Bodén
School of Chemistry and Molecular Biosciences, The University of Queensland
Contact: Jhih-Siang Lai
Abstract
<html>
<p class=MsoNormal style='text-align:justify;text-justify:inter-ideograph'><a
name="_GoBack"></a><span lang=EN-GB>Ancestral sequence reconstruction has had
recent success in decoding the origins and the determinants of complex protein
functions. However, attempts to reconstruct ancient proteins and phylogenetic
analyses of remote homologues must handle extreme amino-acid sequence diversity
resulting from extended periods of evolutionary change [2]. We exploited the wealth
of protein structures in the Protein Data Bank (PDB) to develop an evolutionary
model based on protein secondary structure. The approach follows the
differences between discrete secondary structure states observed in modern
proteins and those hypothesised in their immediate ancestors [1]. Based on this new
evolutionary model, we implemented maximum likelihood-based phylogenetic
inference tools to reconstruct ancestral secondary structure. The predictive
accuracy from the use of the evolutionary model surpasses that of structure
comparative modelling and sequence-based prediction methods; the reconstruction
extracts information not available from modern structures or the ancestral
protein sequences alone [3,4,5]. Based on a phylogenetic analysis of a sequence-diverse
protein family, we showed that the model has the capacity to highlight
relationships that are evolutionarily rooted in structure and not evident in
sequence-based phylogenetic analysis.</span></p>
</html>
Supplementary Data
Supplementary data for "Evolutionary model of protein secondary structure capable of revealing new biological relationships" can be downloaded from
Dataset (140.3 MB) or Software (617 KB), separately.
Package may be downloaded as a single archive from
Full package download (140.9 MB).
- (Ancestral secondary structure reconstruction) ASSR needs version MATLAB 8.2 (R2013b) or later.
- Please send feedback and questions to author.
References
<html>
<body bgcolor=white lang=ZH-TW link=blue vlink="#954F72" style='tab-interval: 24.0pt;text-justify-trim:punctuation'>
<div class=WordSection1 style='layout-grid:20.0pt'>
<p class=MsoListParagraph style='margin-left:18.0pt;mso-para-margin-left:0gd; text-align:justify;text-justify:inter-ideograph;text-indent:-18.0pt;mso-list: l0 level1 lfo1'><![if !supportLists]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:Helvetica; mso-bidi-font-family:Helvetica;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'><span style='mso-list:Ignore'>1.<span style='font:7.0pt "Times New Roman"'> </span></span></span><![endif]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:"Times New Roman"; mso-bidi-font-family:Arial;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'>Dayhoff M, Schwartz R and <span class=SpellE>Orcutt</span> B (1978), <i>"A Model of Evolutionary Change in Proteins"</i>, In Atlas of protein sequence and structure. Vol. 5, pp. 345-352. National Biomedical Research Foundation Silver Spring, MD.<o:p></o:p></span></p>
<p class=MsoListParagraph style='margin-left:18.0pt;mso-para-margin-left:0gd; text-align:justify;text-justify:inter-ideograph;text-indent:-18.0pt;mso-list: l0 level1 lfo1'><![if !supportLists]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:Helvetica; mso-bidi-font-family:Helvetica;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'><span style='mso-list:Ignore'>2.<span style='font:7.0pt "Times New Roman"'> </span></span></span><![endif]><span class=SpellE><span lang=EN-US style='font-size:10.0pt;font-family:"Helvetica",sans-serif;mso-fareast-font-family: "Times New Roman";mso-bidi-font-family:Arial;color:#2E2E2E;mso-font-kerning: 0pt;mso-ansi-language:EN-US'>Ve</span></span><span lang=EN-US style='font-size: 10.0pt;font-family:"Helvetica",sans-serif;mso-fareast-font-family:"Times New Roman"; mso-bidi-font-family:Arial;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'> T, Williams SJ and Kobe B (2015), <i>"Structure and function of Toll/interleukin-1 receptor/resistance protein (TIR) domains"</i>, Apoptosis. Vol. 20(2), pp. 250-261.<o:p></o:p></span></p>
<p class=MsoListParagraph style='margin-left:18.0pt;mso-para-margin-left:0gd; text-align:justify;text-justify:inter-ideograph;text-indent:-18.0pt;mso-list: l0 level1 lfo1'><![if !supportLists]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:Helvetica; mso-bidi-font-family:Helvetica;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'><span style='mso-list:Ignore'>3.<span style='font:7.0pt "Times New Roman"'> </span></span></span><![endif]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:"Times New Roman"; mso-bidi-font-family:Arial;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'>Clifton BE and Jackson CJ (2016), <i>"Ancestral Protein Reconstruction Yields Insights into Adaptive Evolution of Binding Specificity in Solute-Binding Proteins"</i>, Cell <span class=SpellE>Chem</span> Biol. Vol. 23(2), pp. 236-245.<o:p></o:p></span></p>
<p class=MsoListParagraph style='margin-left:18.0pt;mso-para-margin-left:0gd; text-align:justify;text-justify:inter-ideograph;text-indent:-18.0pt;mso-list: l0 level1 lfo1'><![if !supportLists]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:Helvetica; mso-bidi-font-family:Helvetica;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'><span style='mso-list:Ignore'>4.<span style='font:7.0pt "Times New Roman"'> </span></span></span><![endif]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:"Times New Roman"; mso-bidi-font-family:Arial;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'>Hudson WH, <span class=SpellE>Kossmann</span> BR, de Vera IM, Chuo SW, <span class=SpellE>Weikum</span> ER, <span class=SpellE>Eick</span> GN, Thornton JW, Ivanov IN, <span class=SpellE>Kojetin</span> DJ and <span class=SpellE>Ortlund</span> EA (2016), <i>"Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space"</i>, Proc Natl <span class=SpellE>Acad</span> <span class=SpellE>Sci</span> U S A. Vol. 113(2), pp. 326-331.<o:p></o:p></span></p>
<p class=MsoListParagraph style='margin-left:18.0pt;mso-para-margin-left:0gd; text-align:justify;text-justify:inter-ideograph;text-indent:-18.0pt;mso-list: l0 level1 lfo1'><![if !supportLists]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:Helvetica; mso-bidi-font-family:Helvetica;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'><span style='mso-list:Ignore'>5.<span style='font:7.0pt "Times New Roman"'> </span></span></span><![endif]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:"Times New Roman"; mso-bidi-font-family:Arial;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'>Wilson C, <span class=SpellE>Agafonov</span> RV, <span class=SpellE>Hoemberger</span> M, <span class=SpellE>Kutter</span> S, Zorba A, <span class=SpellE>Halpin</span> J, <span class=SpellE>Buosi</span> V, <span class=SpellE>Otten</span> R, Waterman D, Theobald DL and Kern D (2015), <i>"Using ancient protein kinases to unravel a modern cancer drug's mechanism"</i>, Science. Vol. 347(6224), pp. 882-886.<o:p></o:p></span></p>
</div>
</body>
</html>
Application
The Evolu-sec was applied for analysing the structural divergence of Toll/interleukin-1 receptor domains further including plant RUN1 TIR domain and human SARM1 TIR domain, published in <html> <head> <meta http-equiv=Content-Type content="text/html; charset=utf-8"> <style> <!– @font-face
{font-family:Helvetica;
panose-1:0 0 0 0 0 0 0 0 0 0;}
@font-face
{font-family:"Cambria Math";
panose-1:2 4 5 3 5 4 6 3 2 4;}
p.MsoNormal, li.MsoNormal, div.MsoNormal
{margin:0cm;
margin-bottom:.0001pt;
font-size:10.0pt;
font-family:Helvetica;}
a:link, span.MsoHyperlink
{color:#0563C1;
text-decoration:underline;}
.MsoChpDefault
{font-family:Helvetica;}
–> </style>
</head>
<body lang=EN-AU link="#0563C1" vlink="#954F72">
<div class=WordSection1>
<p class=MsoNormal><a name="_GoBack"></a>"NAD+ cleavage activity by animal and plant TIR domains in cell death pathways, <i>Science</i>, 365, 6455, 2019."</p>
<p class=MsoNormal><span lang=EN-GB><a href="http://www.doi.org/10.1126/science.aax1911">http://www.doi.org/10.1126/science.aax1911</a></span></p>
<p class=MsoNormal><span lang=EN-GB> </span></p>
</div> </body> </html> This wiki page provides images of better resolution, which have been included in the supplementary dataset of "NAD+ cleavage activity by animal and plant TIR domains in cell death pathways".
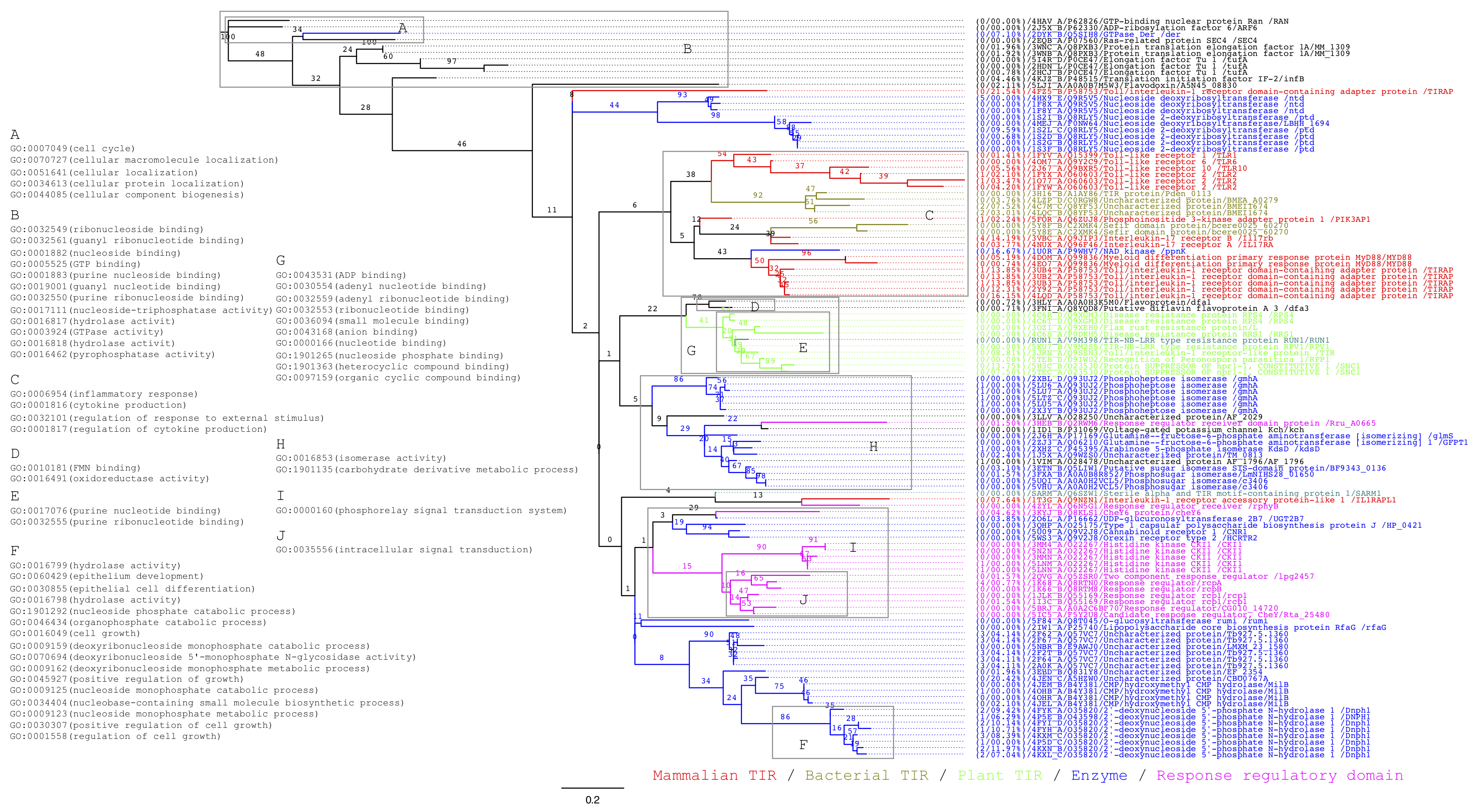 Figure 1. Unrooted phylogenetic tree of proteins containing TIR domains and structurally related domains. The phylogenetic tree includes 114 proteins from the intersection of DALI searches on human SARM1 TIR and plant RUN1 TIR. Only non-redundant proteins with structures at 4Å resolution or better and superpositions covering 125 residues or more in length were used. Numbers on branches are standard bootstrap values by resampling 100 times. Sub-trees are coloured to indicate kingdom and broad function and annotated with Gene Ontology terms that are statistically enriched for the group of proteins at the leaves. Each leaf specifies PDB and UniProt identifiers, protein and gene names; in addition, the absolute number of mutated amino acids and the ratio of missing residues in the structure relative to the protein sequence are provided.
Figure 1. Unrooted phylogenetic tree of proteins containing TIR domains and structurally related domains. The phylogenetic tree includes 114 proteins from the intersection of DALI searches on human SARM1 TIR and plant RUN1 TIR. Only non-redundant proteins with structures at 4Å resolution or better and superpositions covering 125 residues or more in length were used. Numbers on branches are standard bootstrap values by resampling 100 times. Sub-trees are coloured to indicate kingdom and broad function and annotated with Gene Ontology terms that are statistically enriched for the group of proteins at the leaves. Each leaf specifies PDB and UniProt identifiers, protein and gene names; in addition, the absolute number of mutated amino acids and the ratio of missing residues in the structure relative to the protein sequence are provided.
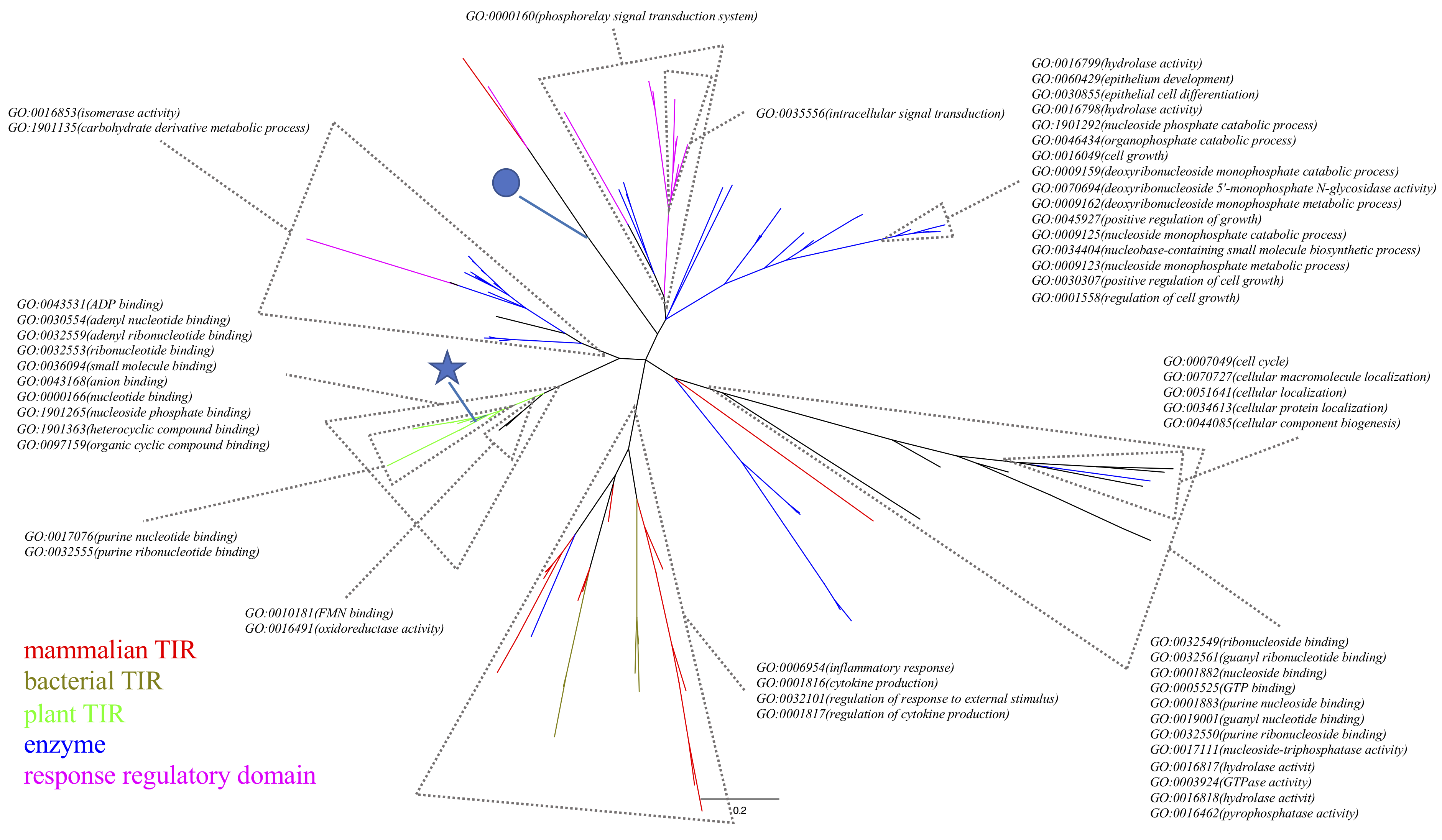 Figure 2. The alternative tree visualisation of Figure 1. Tips in red, brown, green, blue, purple are mammalian TIR domains, bacterial TIR domains, plant TIR domains, enzymes, and response regulatory domains, respectively. The star and the circle highlight the plant RUN1 TIR and human SARM1 TIR structures, respectively.
Figure 2. The alternative tree visualisation of Figure 1. Tips in red, brown, green, blue, purple are mammalian TIR domains, bacterial TIR domains, plant TIR domains, enzymes, and response regulatory domains, respectively. The star and the circle highlight the plant RUN1 TIR and human SARM1 TIR structures, respectively.
External links
Authors
Created by admin (Administrator) on 2018/07/11 12:44.
Contributing authors: