This is an old revision of the document!
Evo-SS-Package
Evo-SS-Package is a part of research project as “Modelling the evolution of protein secondary structure”.
Evo-SS-Package contains phylogenetic analysis tools based on an evolutionary model of secondary structure.
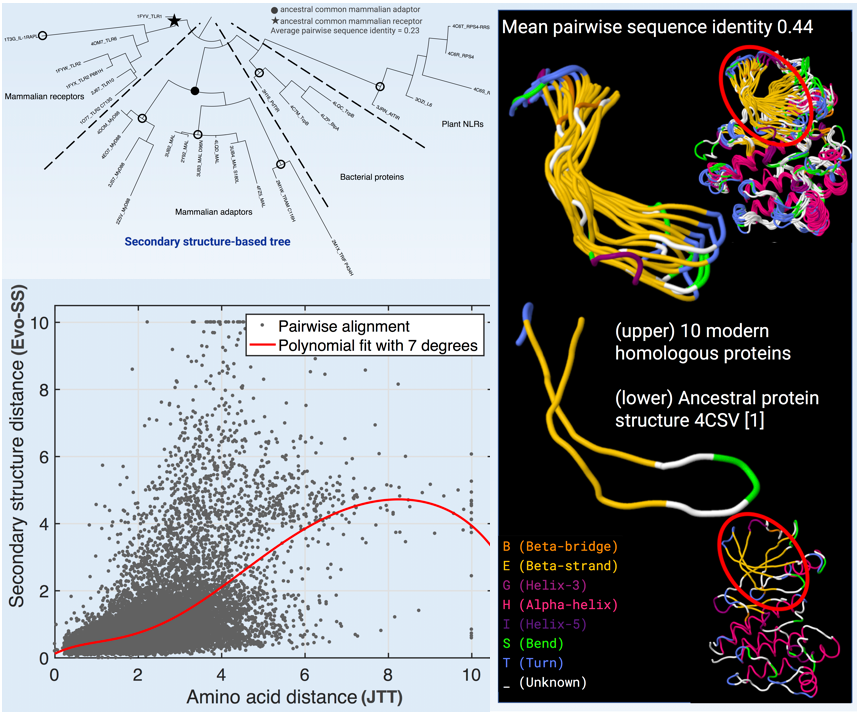
Analysis focus on phylogenetic tree inference [left-top], evolutionary distance estimation [left-bottom], and ancestral secondary structure reconstruction [right].
The research project abstract is provided in next session.
The dataset for the model development and the software can be accessed in the “Supplementary Data” session.
Research Project
Modelling the evolution of protein secondary structure
Jhih-Siang Lai, Bostjan Kobe and Mikael Bodén
School of Chemistry and Molecular Biosciences, The University of Queensland
Contact: Jhih-Siang Lai
Abstract
Ancestral sequence reconstruction has had recent success in decoding the origins and the determinants of complex protein functions. However, attempts to reconstruct extremely ancient proteins and phylogenetic analyses of remote homologues must deal with the sequence diversity that results from extended periods of evolutionary change. In the last twenty years, the number of protein structures in the Protein Data Bank has increased twenty-fold. Using the same principles pioneered by Dayhoff [1], we seize this wealth of structure data and develop a protein secondary structure evolutionary model, based on differences between discrete secondary structure states observed in modern proteins and those hypothesized in their immediate ancestors. We implement maximum likelihood-based phylogenetic inference tools based on our evolutionary model. We apply these tools to the sequence-diverse but structurally-conserved Toll/interleukin-1 receptor (TIR) domains [2] and show that the resulting clades in a phylogenetic tree are more consistent with their biological properties than those of the same inference based on an amino acid model. The approach also allows us to infer ancestral secondary structure [3,4,5]; we compare these predictions with those of structure homology modelling and sequence-based secondary structure predictors. The secondary structure evolutionary model extracts information not available from modern structures or the ancestral protein sequences alone. Our evolutionary model has the capacity to highlight relationships that are evolutionarily rooted in structure, and therefore complements the use of sequence-based phylogenetic analysis.
Supplementary Data
Supplementary data for "Modelling the evolution of protein secondary structure" can be downloaded as
Dataset or Software, separately.
Package may be downloaded as single one archive as
Full package download.
Package contents can be viewed here.
Supplementary document
- (Ancestral secondary structure reconstruction) ASSR needs version MATLAB 8.2 (R2013b) or later.
- Please feedback needs to the author.
References
<html>
<body bgcolor=white lang=ZH-TW link=blue vlink="#954F72" style='tab-interval: 24.0pt;text-justify-trim:punctuation'>
<div class=WordSection1 style='layout-grid:20.0pt'>
<p class=MsoListParagraph style='margin-left:18.0pt;mso-para-margin-left:0gd; text-align:justify;text-justify:inter-ideograph;text-indent:-18.0pt;mso-list: l0 level1 lfo1'><![if !supportLists]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:Helvetica; mso-bidi-font-family:Helvetica;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'><span style='mso-list:Ignore'>1.<span style='font:7.0pt "Times New Roman"'> </span></span></span><![endif]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:"Times New Roman"; mso-bidi-font-family:Arial;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'>Dayhoff M, Schwartz R and <span class=SpellE>Orcutt</span> B (1978), <i>"A Model of Evolutionary Change in Proteins"</i>, In Atlas of protein sequence and structure. Vol. 5, pp. 345-352. National Biomedical Research Foundation Silver Spring, MD.<o:p></o:p></span></p>
<p class=MsoListParagraph style='margin-left:18.0pt;mso-para-margin-left:0gd; text-align:justify;text-justify:inter-ideograph;text-indent:-18.0pt;mso-list: l0 level1 lfo1'><![if !supportLists]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:Helvetica; mso-bidi-font-family:Helvetica;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'><span style='mso-list:Ignore'>2.<span style='font:7.0pt "Times New Roman"'> </span></span></span><![endif]><span class=SpellE><span lang=EN-US style='font-size:10.0pt;font-family:"Helvetica",sans-serif;mso-fareast-font-family: "Times New Roman";mso-bidi-font-family:Arial;color:#2E2E2E;mso-font-kerning: 0pt;mso-ansi-language:EN-US'>Ve</span></span><span lang=EN-US style='font-size: 10.0pt;font-family:"Helvetica",sans-serif;mso-fareast-font-family:"Times New Roman"; mso-bidi-font-family:Arial;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'> T, Williams SJ and Kobe B (2015), <i>"Structure and function of Toll/interleukin-1 receptor/resistance protein (TIR) domains"</i>, Apoptosis. Vol. 20(2), pp. 250-261.<o:p></o:p></span></p>
<p class=MsoListParagraph style='margin-left:18.0pt;mso-para-margin-left:0gd; text-align:justify;text-justify:inter-ideograph;text-indent:-18.0pt;mso-list: l0 level1 lfo1'><![if !supportLists]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:Helvetica; mso-bidi-font-family:Helvetica;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'><span style='mso-list:Ignore'>3.<span style='font:7.0pt "Times New Roman"'> </span></span></span><![endif]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:"Times New Roman"; mso-bidi-font-family:Arial;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'>Clifton BE and Jackson CJ (2016), <i>"Ancestral Protein Reconstruction Yields Insights into Adaptive Evolution of Binding Specificity in Solute-Binding Proteins"</i>, Cell <span class=SpellE>Chem</span> Biol. Vol. 23(2), pp. 236-245.<o:p></o:p></span></p>
<p class=MsoListParagraph style='margin-left:18.0pt;mso-para-margin-left:0gd; text-align:justify;text-justify:inter-ideograph;text-indent:-18.0pt;mso-list: l0 level1 lfo1'><![if !supportLists]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:Helvetica; mso-bidi-font-family:Helvetica;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'><span style='mso-list:Ignore'>4.<span style='font:7.0pt "Times New Roman"'> </span></span></span><![endif]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:"Times New Roman"; mso-bidi-font-family:Arial;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'>Hudson WH, <span class=SpellE>Kossmann</span> BR, de Vera IM, Chuo SW, <span class=SpellE>Weikum</span> ER, <span class=SpellE>Eick</span> GN, Thornton JW, Ivanov IN, <span class=SpellE>Kojetin</span> DJ and <span class=SpellE>Ortlund</span> EA (2016), <i>"Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space"</i>, Proc Natl <span class=SpellE>Acad</span> <span class=SpellE>Sci</span> U S A. Vol. 113(2), pp. 326-331.<o:p></o:p></span></p>
<p class=MsoListParagraph style='margin-left:18.0pt;mso-para-margin-left:0gd; text-align:justify;text-justify:inter-ideograph;text-indent:-18.0pt;mso-list: l0 level1 lfo1'><![if !supportLists]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:Helvetica; mso-bidi-font-family:Helvetica;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'><span style='mso-list:Ignore'>5.<span style='font:7.0pt "Times New Roman"'> </span></span></span><![endif]><span lang=EN-US style='font-size:10.0pt; font-family:"Helvetica",sans-serif;mso-fareast-font-family:"Times New Roman"; mso-bidi-font-family:Arial;color:#2E2E2E;mso-font-kerning:0pt;mso-ansi-language: EN-US'>Wilson C, <span class=SpellE>Agafonov</span> RV, <span class=SpellE>Hoemberger</span> M, <span class=SpellE>Kutter</span> S, Zorba A, <span class=SpellE>Halpin</span> J, <span class=SpellE>Buosi</span> V, <span class=SpellE>Otten</span> R, Waterman D, Theobald DL and Kern D (2015), <i>"Using ancient protein kinases to unravel a modern cancer drug's mechanism"</i>, Science. Vol. 347(6224), pp. 882-886.<o:p></o:p></span></p>
</div>
</body>
</html>
External links
Sean in ResearchGate
Sean's profile
Last updated by — Jhih-Siang Lai 2017/07/11 21:53